CBIOMES
Constraint-based modeling of marine microbial community metabolism and physiology
As part of the CBIOMES Project, we are designing detailed models of microbial metabolism and physiology to simulate microbial dynamics in the marine environment from a mechanistic, bottom-up perspective.
Two MATLAB toolboxes are under development for this project: The Microbiome Simulation Environment toolbox (MSE) and the PanGEnome-scale Metabolic model toolbox PanGEM. Lots more on them in time!
Here’s a primer on what’s to come…
And here’s a more recent summary of our application of MSE to an Atlantic Meridional Transect cruise…
Mesoscale dynamics of Prochlorococcus
Update: Our first full-scale simulation – Prochlorococcus growth across an eddy dipole transect – is complete! We are thrilled with the results and are currently sifting through all the data to develop a compelling story.
The model was run using in situ data collected on the 2017 MESO-SCOPE Cruise, including depth profiles of flow cytometry cell counts and scattering properties, ITS-based relative abundances, nutrient concentrations, spectral irradiance, and temperature. These data are fed at each grid point to genome-scale metabolic models (GEMs) for 48 sequenced isolate strains. Each GEM goes through a series of bilevel optimizations which allow the cell to physiologically acclimate to its environment by altering its size, its macromolecular composition, its pigment composition, and the abundance and types of transporters on the cell surface. Using the acclimated GEM and the associated constraints, flux balance analysis is then run to predict growth rates and metabolic fluxes. Finally, we apply a modified Arrhenius function using optimal growth temperatures derived from a machine learning algorithm. Here is a teaser:
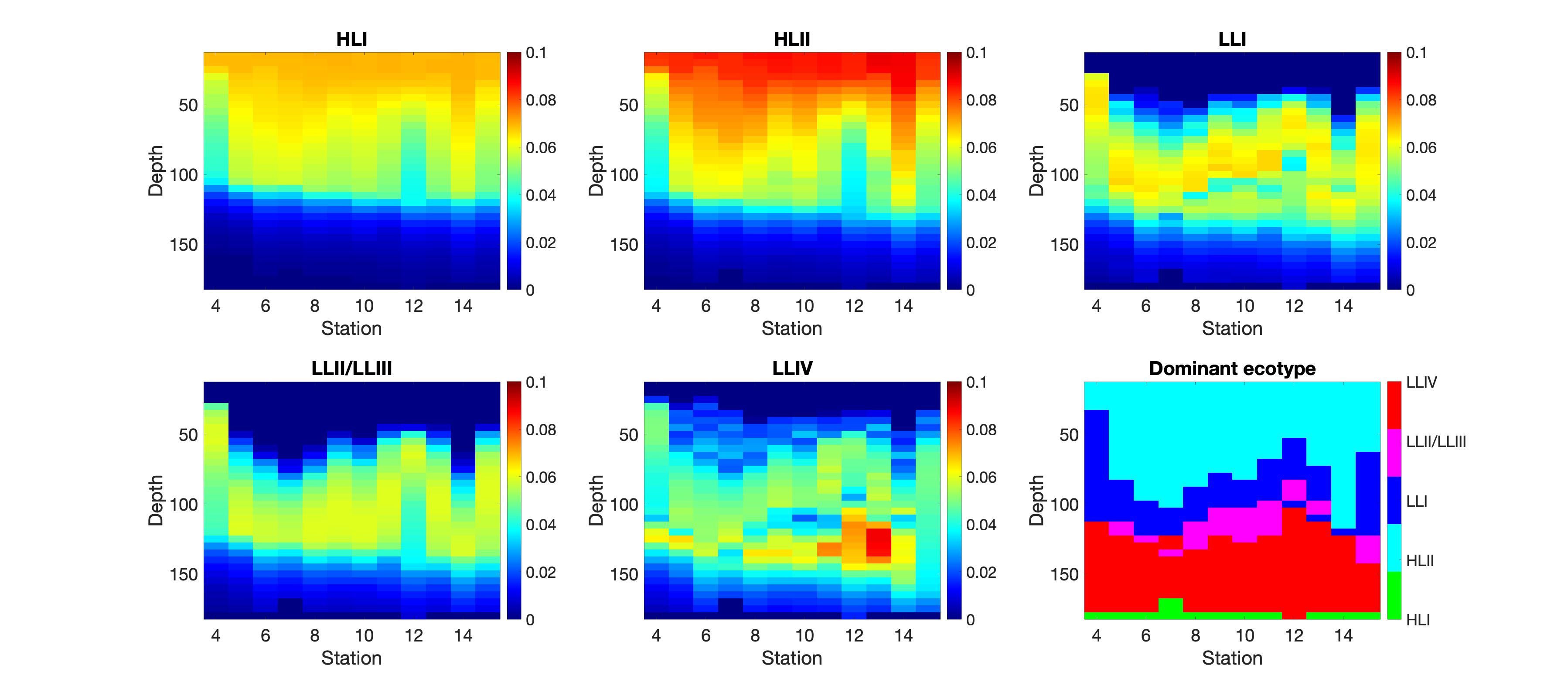
Ecotype averaged growth rates (h-1), predicted at local noon across the MESO-SCOPE transect. Station 6 is the center of the anti-cyclone; Station 12 is the center of the cyclone. 5 Ecotypes are shown: High-Light adapted ecotypes I and II, and Low-Light adapted ecotypes I II/III and IV. Don’t worry about that HLI at the bottom lol.
There seems to be either a boundless flow of genes or a vast amount of persistent micro-diversity within the Prochlorococcus community. We could never hope to capture all the instances in GEMs, so instead we are evolving in silico strains synthesized from the pangenome using our PanGEM Toolbox. In addition to the 48 isolates, the pangenome also includes genes from 97 strains built from MAGs and SAGs, and we plan to include an additional 489 SAGs recently published by Berube et al., 2018.
